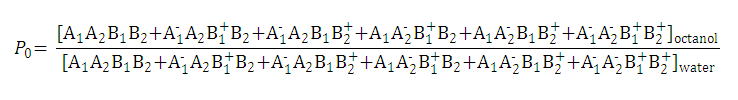
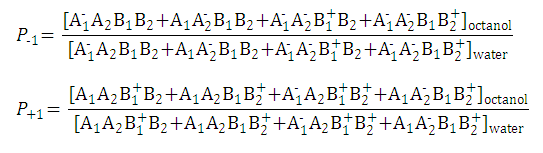
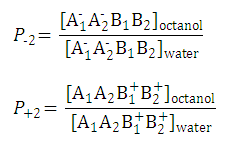
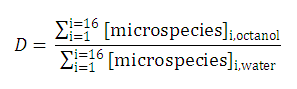

| 1. Partition coefficient of the neutral species: |
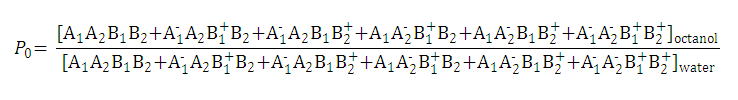
| |||
| 2. Partition coefficient of the anionic and the cationic species: |
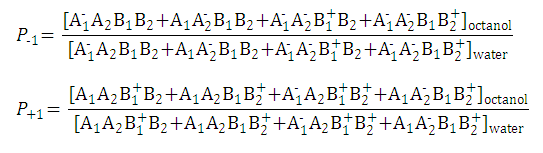
|
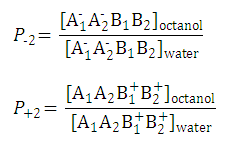
| ||
| 3. The distribution coefficient of the A1A2B1B2 molecule: | 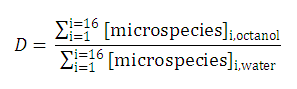 | |||

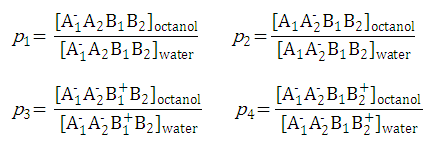
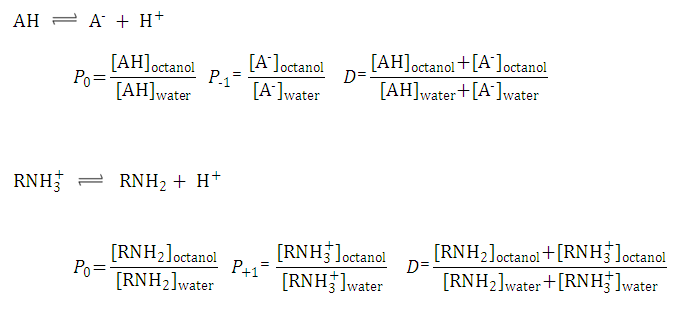
After substituting the pis into the original formula for P-1 we get the following simpler formula which includes only aqueous concentration of the appropriate microspecies:

This can be further simplified if we introduce the acid dissociation constants of the A1A2B1B2 molecule. The next five ionization reactions of the A1A2B1B2 molecule are used to rearrange P-1 into a concentration free form.
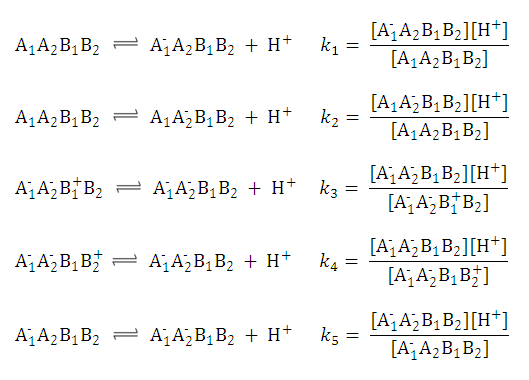

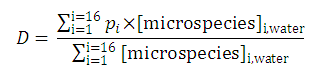
logP calculations are based on a pool of fragments predefined
in the calculator. This set is based on the data set in
references 1. Every fragment is assigned a unique name and a value. |
|
The logP value of a molecule
is the sum of the fragment values present in the molecule. |
|
logP plugin handled
only one fragment set until version 5.1.2, above, it was extended with two additional
sets. The sets are based on a published data set (see
reference 2) and the PhysProp© database. |
| The trainable logP (available from version 5.1.3) offers the user to define
his own logP database and calculate logP values based on the experimental data.
New fragment value extensions make a more precise calculation possible. |
| Choosing the weighted method requires the user to define the weight of each
set of data needed. This method may include any of the three internal data sets and
the user defined set. |
 |
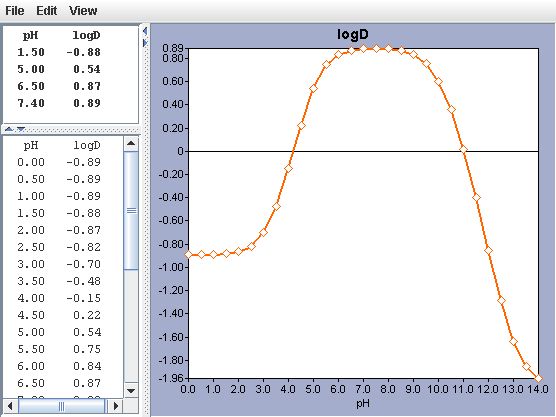 |
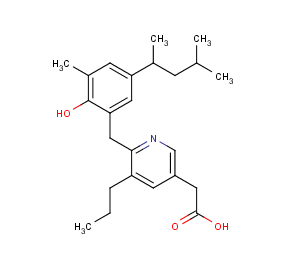
| Calculated logP = -1.09 | measured logP= -1.10 | , see Ref.3. |